-
Lukáš Krupčík authoredLukáš Krupčík authored
Bioinformatics Applications
Introduction
In addition to the many applications available through modules (deployed through EasyBuild packaging system) we provide an alternative source of applications on our clusters inferred from Gentoo Linux. The user's environment is setup through a script which returns a bash instance to the user (you can think of it a starting a whole virtual machine but inside your current namespace) . The applications were optimized by gcc compiler for the SandyBridge and IvyBridge platforms. The binaries use paths from /apps/gentoo prefix to find the required runtime dependencies, config files, etc. The Gentoo Linux is a standalone installation not even relying on the glibc provided by host operating system (Redhat). The trick which allowed us to install Gentoo Linux on the host Redhat system is called Gentoo::RAP and uses a modified loader with a hardcoded path (links).
Starting the Environment
mmokrejs@login2~$ /apps/gentoo/startprefixStarting PBS Jobs Using the Applications
Create a template file which can be used and an argument to qsub command. Notably, the 'PBS -S' line specifies full PATH to the Bourne shell of the Gentoo Linux environment.
mmokrejs@login2~$ cat myjob.pbs
#PBS -S /apps/gentoo/bin/sh
#PBS -l nodes=1:ppn=16,walltime=12:00:00
#PBS -q qfree
#PBS -M my_email@foo.bar
#PBS -m ea
#PBS -N sample22
#PBS -A DD-13-5
#source ~/.bashrc
cd $PBS_O_WORKDIR || exit 255
myscript.sh foo 1>myjob.log 2>&1
$ head -n 1 myscript.sh
#! /apps/gentoo/bin/sh
$ qsub myjob.pbs
$ qstatReading Manual Pages for Installed Applications
mmokrejs@login2~$ man -M /apps/gentoo/usr/share/man bwa
mmokrejs@login2~$ man -M /apps/gentoo/usr/share/man samtoolsListing of Bioinformatics Applications
mmokrejs@login2~$ grep biology /scratch/mmokrejs/gentoo_rap/installed.txt
sci-biology/ANGLE-bin-20080813-r1
sci-biology/AlignGraph-9999
sci-biology/Atlas-Link-0.01-r1
sci-biology/BRANCH-9999
sci-biology/EBARDenovo-1.2.2
sci-biology/FLASH-1.2.9
sci-biology/GAL-0.2.2
sci-biology/Gambit-0.4.145
sci-biology/HTSeq-0.6.1
sci-biology/InterMine-0.98
sci-biology/MochiView-1.45
sci-biology/MuSeqBox-5.4
sci-biology/ONTO-PERL-1.41
sci-biology/ORFcor-20130507
sci-biology/Rcorrector-9999
sci-biology/SSAKE-3.8.2
sci-biology/STAR-9999
sci-biology/YASRA-2.33
sci-biology/abacas-1.3.1
sci-biology/align_to_scf-1.06
sci-biology/assembly-stats-9999
sci-biology/bambus-2.33
sci-biology/bamtools-9999
sci-biology/bcftools-1.2
sci-biology/bedtools-2.22.1
sci-biology/bfast-0.7.0a
sci-biology/biobambam2-9999
sci-biology/bismark-0.13.0
sci-biology/blat-34-r1
sci-biology/blue-1.1.3
sci-biology/bowtie-2.2.9
sci-biology/brat-1.2.4
sci-biology/bwa-0.7.13
sci-biology/bx-python-9999
sci-biology/cast-bin-20080813
sci-biology/cd-hit-4.6.5
sci-biology/cdbfasta-0.1
sci-biology/clover-2011.10.24
sci-biology/clustalw-2.1
sci-biology/cnrun-2.0.3
sci-biology/codonw-1.4.4-r2
sci-biology/conform-gt-1174
sci-biology/conifer-0.2.2
sci-biology/coral-1.4
sci-biology/cross_genome-20140822
sci-biology/cutadapt-9999
sci-biology/dawg-1.1.2
sci-biology/dna2pep-1.1
sci-biology/edena-3.131028
sci-biology/epga-9999
sci-biology/erpin-5.5b
sci-biology/estscan-3.0.3
sci-biology/eugene-4.1d
sci-biology/exonerate-gff3-9999
sci-biology/fastx_toolkit-0.0.14
sci-biology/gemini-9999
sci-biology/geneid-1.4.4
sci-biology/genepop-4.2.1
sci-biology/glimmerhmm-3.0.1-r1
sci-biology/gmap-2015.12.31.5
sci-biology/hexamer-19990330
sci-biology/hts-python-9999
sci-biology/jellyfish-2.1.4
sci-biology/jigsaw-3.2.10
sci-biology/kallisto-9999
sci-biology/karect-1.0.0
sci-biology/lastz-1.03.66
sci-biology/libgtextutils-0.6.1
sci-biology/lucy-1.20
sci-biology/megahit-9999
sci-biology/merlin-1.1.2
sci-biology/miranda-3.3a
sci-biology/mreps-2.5
sci-biology/mrfast-2.6.0.1
sci-biology/mummer-3.22-r1
sci-biology/muscle-3.8.31
sci-biology/nrcl-110625
sci-biology/nwalign-0.3.1
sci-biology/oases-9999
sci-biology/parafly-20130121
sci-biology/phrap-1.080812-r1
sci-biology/phred-071220
sci-biology/phylip-3.696-r1
sci-biology/plinkseq-0.10
sci-biology/primer3-2.3.7
sci-biology/prinseq-lite-0.20.4
sci-biology/proda-1.0
sci-biology/pybedtools-0.6.9
sci-biology/pysam-0.9.0
sci-biology/pysamstats-0.24.2
sci-biology/quast-2.3
sci-biology/quorum-1.0.0
sci-biology/reaper-15348
sci-biology/repeatmasker-libraries-20150807
sci-biology/reptile-1.1
sci-biology/samstat-20130708
sci-biology/samtools-0.1.20-r2
sci-biology/samtools-1.3-r1
sci-biology/scaffold_builder-20131122-r1
sci-biology/scan_for_matches-20121220
sci-biology/screed-0.7.1
sci-biology/scythe-0.992
sci-biology/seqan-2.1.1
sci-biology/seqtools-4.34.5
sci-biology/sff_dump-1.04
sci-biology/sgp2-1.1
sci-biology/shrimp-2.2.3
sci-biology/sickle-9999
sci-biology/smalt-0.7.6
sci-biology/snpomatic-9999
sci-biology/ssaha2-bin-2.5.5
sci-biology/stampy-1.0.28
sci-biology/stringtie-1.2.2
sci-biology/subread-1.4.6
sci-biology/swissknife-1.72
sci-biology/tagdust-20101028
sci-biology/tclust-110625
sci-biology/tigr-foundation-libs-2.0-r1
sci-biology/trans-abyss-1.4.8
sci-biology/trf-4.07b
sci-biology/uchime-4.2.40
sci-biology/velvet-1.2.10
sci-biology/velvetk-20120606
sci-biology/zmsort-110625mmokrejs@login2~$ grep sci-libs /scratch/mmokrejs/gentoo_rap/installed.txt
sci-libs/amd-2.3.1
sci-libs/blas-reference-20151113-r1
sci-libs/camd-2.3.1
sci-libs/cbflib-0.9.3.3
sci-libs/ccolamd-2.8.0
sci-libs/cholmod-2.1.2
sci-libs/coinor-cbc-2.8.9
sci-libs/coinor-cgl-0.58.6
sci-libs/coinor-clp-1.15.6-r1
sci-libs/coinor-dylp-1.9.4
sci-libs/coinor-osi-0.106.6
sci-libs/coinor-utils-2.9.11
sci-libs/coinor-vol-1.4.4
sci-libs/colamd-2.8.0
sci-libs/cxsparse-3.1.2
sci-libs/dcmtk-3.6.0
sci-libs/gsl-2.1
sci-libs/hdf5-1.8.15_p1
sci-libs/htslib-1.3
sci-libs/io_lib-1.14.7
sci-libs/lapack-reference-3.6.0-r1
sci-libs/lemon-1.3-r2
sci-libs/libmaus2-9999
sci-libs/qrupdate-1.1.2-r1
sci-libs/scikits-0.1-r1
sci-libs/suitesparseconfig-4.2.1
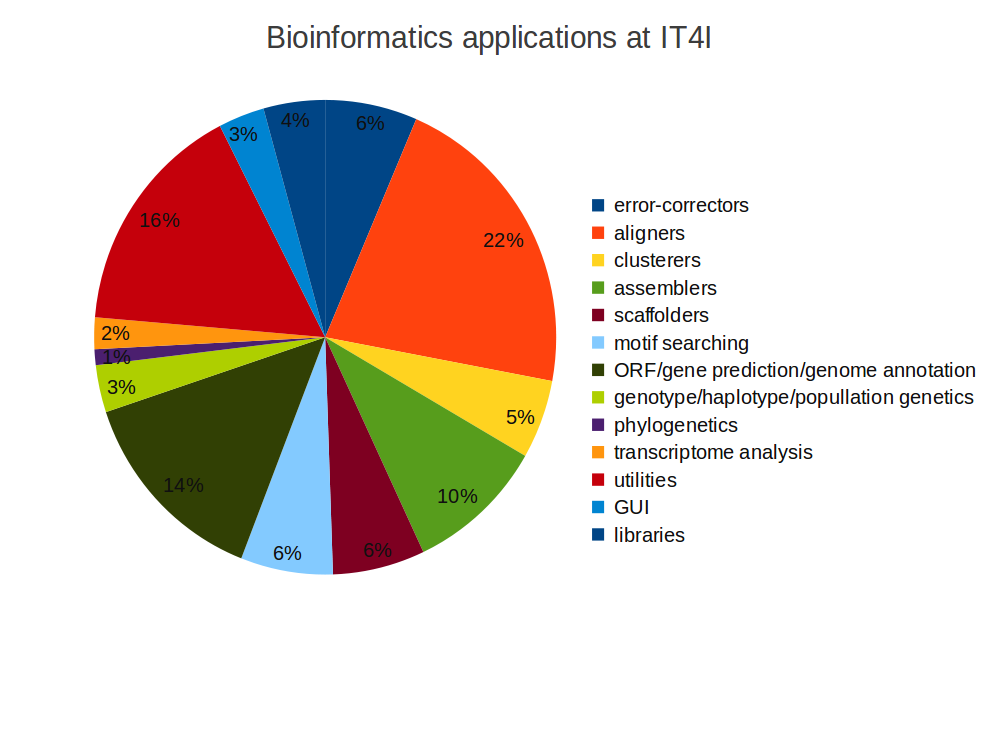
sci-libs/umfpack-5.6.2Classification of Applications
| Applications for bioinformatics at IT4I | |
|---|---|
| error-correctors | 6 |
| aligners | 20 |
| clusterers | 5 |
| assemblers | 9 |
| scaffolders | 6 |
| motif searching | 6 |
| ORF/gene prediction/genome annotation | 13 |
| genotype/haplotype/popullation genetics | 3 |
| phylogenetics | 1 |
| transcriptome analysis | 2 |
| utilities | 15 |
| GUI | 3 |
| libraries | 4 |
| Total | 93 |
Other Applications Available Through Gentoo Linux
Gentoo Linux is a allows compilation of its applications from source code while using compiler and optimize flags set to user's wish. This facilitates creation of optimized binaries for the host platform. Users maybe also use several versions of gcc, python and other tools.
mmokrejs@login2~$ gcc-config -l
mmokrejs@login2~$ java-config -L
mmokrejs@login2~$ eselect